Contents toc¶
Goal¶
Viewing data on the microscope computer is hard for a number of reasons:
-
We are often simultaneously collecting data, and we don't want to crash the computer.
-
This is the only Windows computer while all other computers are Linux.
-
We have to laboriously open each image sequence as a stack and adjust thresholds depending on the channel.
-
Multiple image stacks do not have the same thresholds, so they are not directly visually comparable.
Strategy¶
-
We will transfer the data using
rsyncfrom theWindowscomputer runningUbuntu WSLto our local desktop. Note that this is a temporary location. All microscope data should be transferred to the Google drive ofrasi2@uw.edudirectly from the microscope computer at the end of the experiment. -
We will run an
ImageJpythonscript to:-
open all stacks of interest, channels of interest (each channel will be a separate stack).
-
adjust the lowest and highest displayed pixel value to be the same across all stacks so that they can be compared.
-
place the different stack windows nicely so that they are not on top of each other (works perfectly only for < 10 stacks).
-
Preliminary Setup on microscope computer¶
-
Open
Windows Terminalas an administrator. -
Stop the Windows
sshservice:Stop-Service sshd -
Open a new tab on the same
Windows Terminalwindow but runningUbuntu. To do this, click on the down arrow at the top of the window bar. -
Start the Ubuntu
sshservice:sudo service ssh --full-restart
Procedure¶
You need to do steps 1—4 below only the first time.
-
Clone the
imagej_scriptsto your local computer if you have not done so already, and symbolically link the correct script to your current analysis folder: -
If you have not done it already, make sure to symbolically link
ImageJto yourbinwhich should be in yourPATH: -
Create an
sshprofile on your local computer to enable easy logging into the microscope computer by putting the following at the bottom of your~/.ssh/configfile: -
Enable password-less login to the microscope computer by sending your public key to the scope computer. You need to use the password from here below:
-
Create the correct destination folder in your local desktop to match the data folder on the microscope computer:
-
Transfer the files to your local computer from the microscope computer using
rsync. Here we are transferring the files for just a single imaging position since it can take a while to transfer files. Note the.at the end of the command to indicate that you want files transferred to your current directory. -
Run the
ImageJpythonscript making sure to pick the rightregexpattern for your folders and files. For example, below we want to look at all folders whose names end asPos0,Pos1, …Pos9, and look at only the files inside them that containTxRin their name:
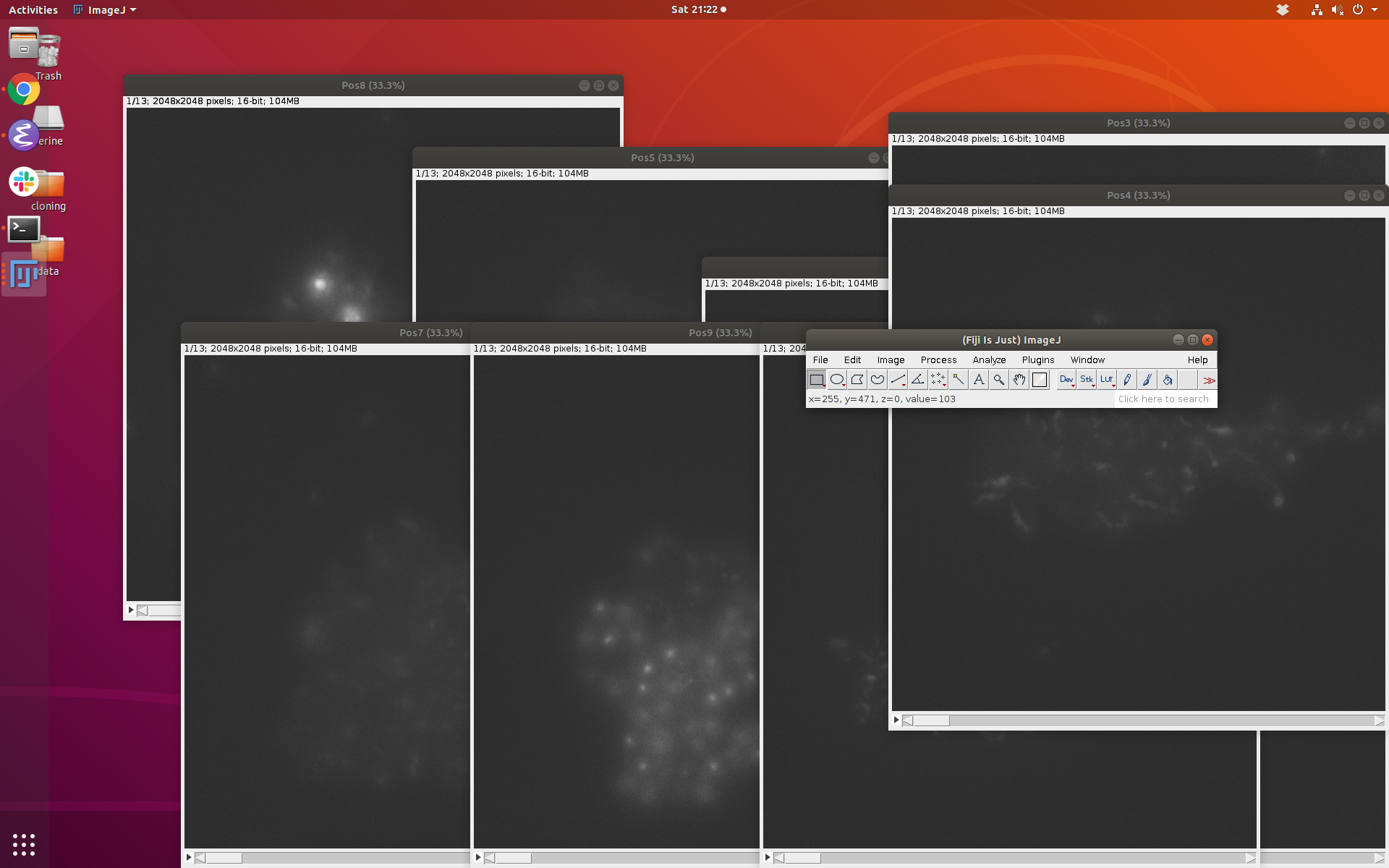
Results¶
Once you run the last line above, you should see ImageJ open up and each of the stacks displayed as a separate window with equal lower and upper thresholds: