Picking restriction sites
If you are designing restriction digests to check inserts, please follow these guidelines to pick enzymes:
- Pick a single (or two) multiple cutter that will cut the final vector 2-4 times instead of several single cutters.
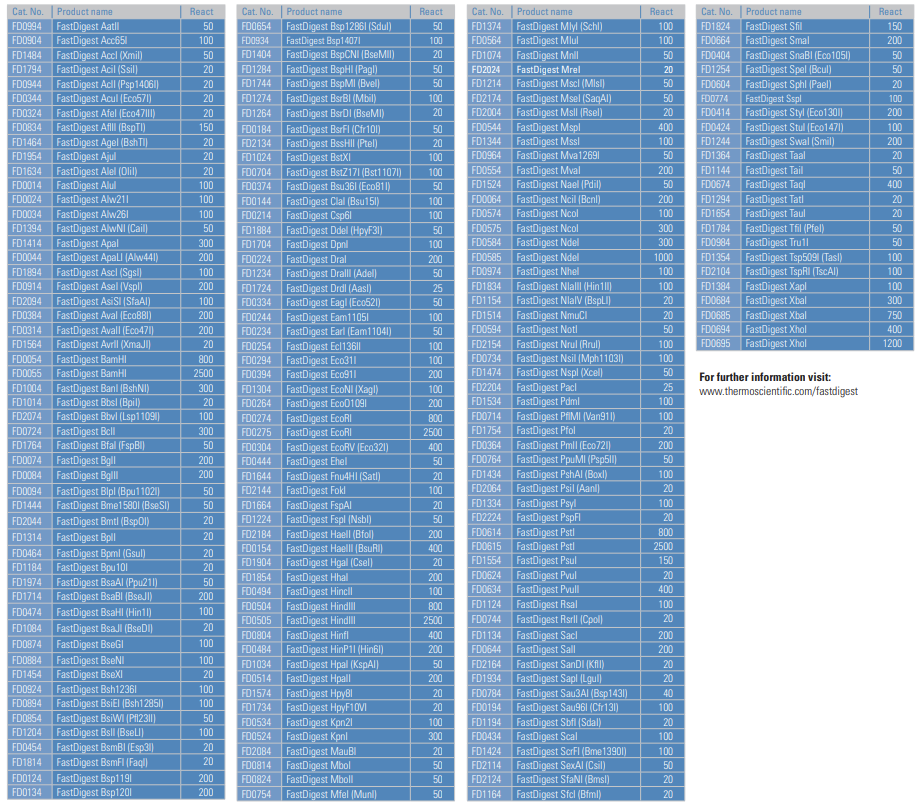
- Pick enzymes that are cheap and available in large quantities (200-800μl). These are also typically the enzymes that cut faster and better. For reference, see the enzymes with higher reaction amounts (>200μl) in image below. Preferred order below. All these enzymes should be in our lab ordering database.
- EcoRI
- EcoRV
- HindIII
- BamHI
- BglII
- XhoI
- PstI
- KpnI
- XbaI
- SalI
- SmaI
- NdeI
- NotI
- NheI
- Pick enzymes that will distinguish between the parent and the cloned vector.
- The digests should give at least one unique band between 1kb and 4kb and this band should be well separated from the remaining cut bands that are common between the parent and the cloned vector. Ideally, all bands will be between 1kb and 4kb, with a subset of bands unique to the cloned vector and the parent vector.
- Do not exchange FastDigest enzymes with non-FastDigest or NEB enzymes. They use different reaction buffers.
- If there is less than 10μl of any of the enzymes you are using, please order more right away.